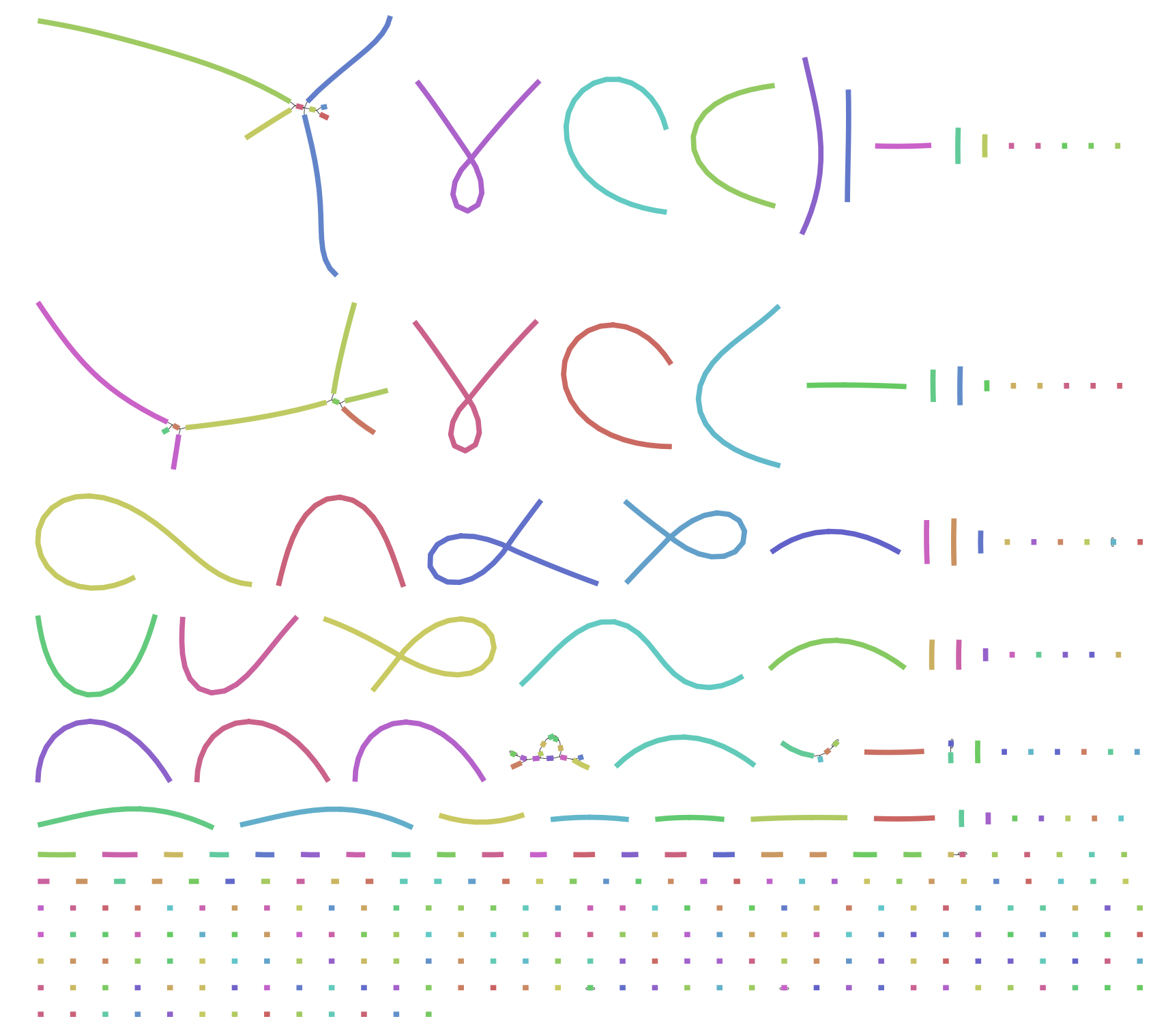Support information
- Category: Genome assembly
- Coordinator: Alexandre Cormier
- Project example: GambOc
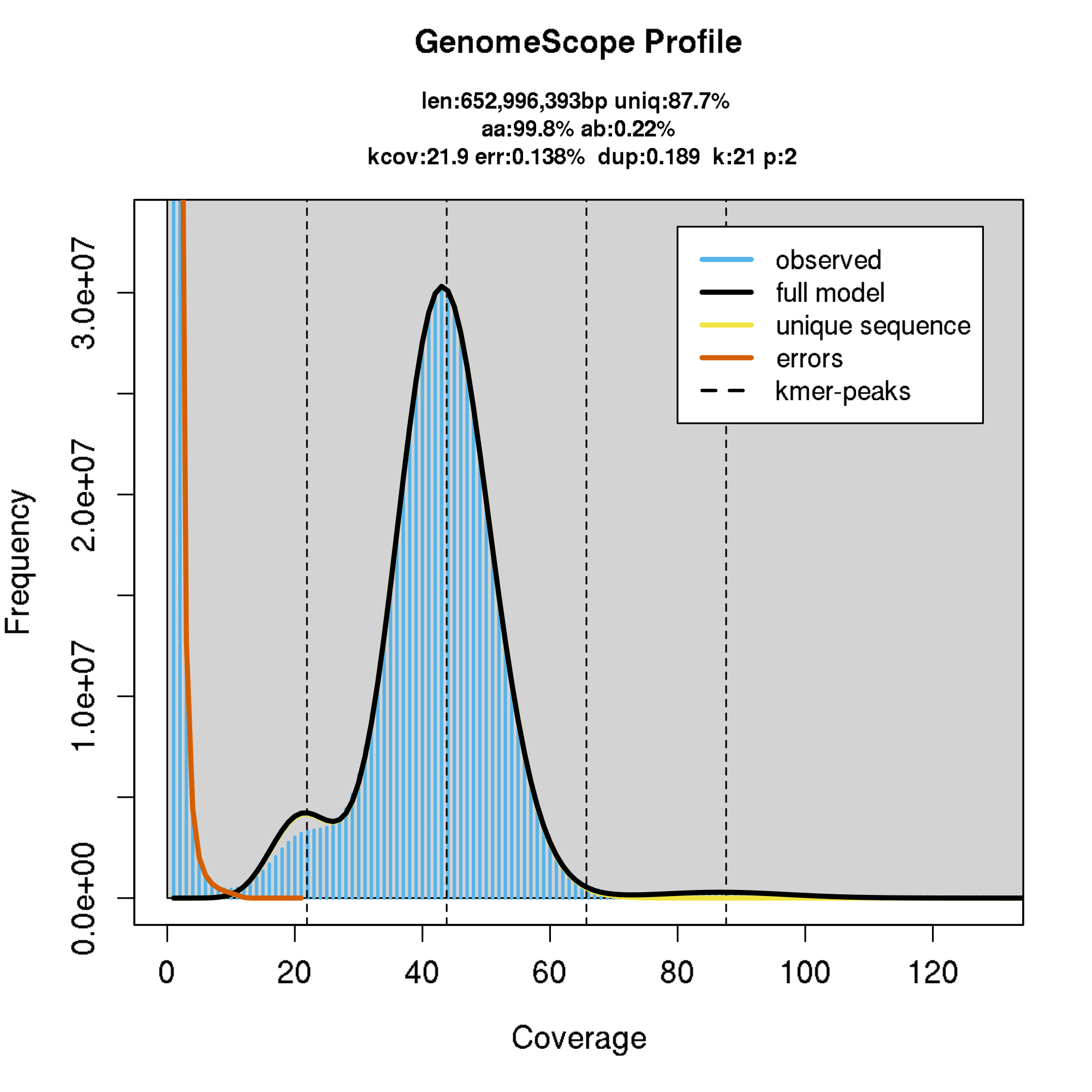
Genome assembly
Recent advances in sequencing technologies, such as PacBio and Oxford Nanopore, have made possible to
sequence and assemble entire genomes in a matter of hours or days with unprecedented quality for a
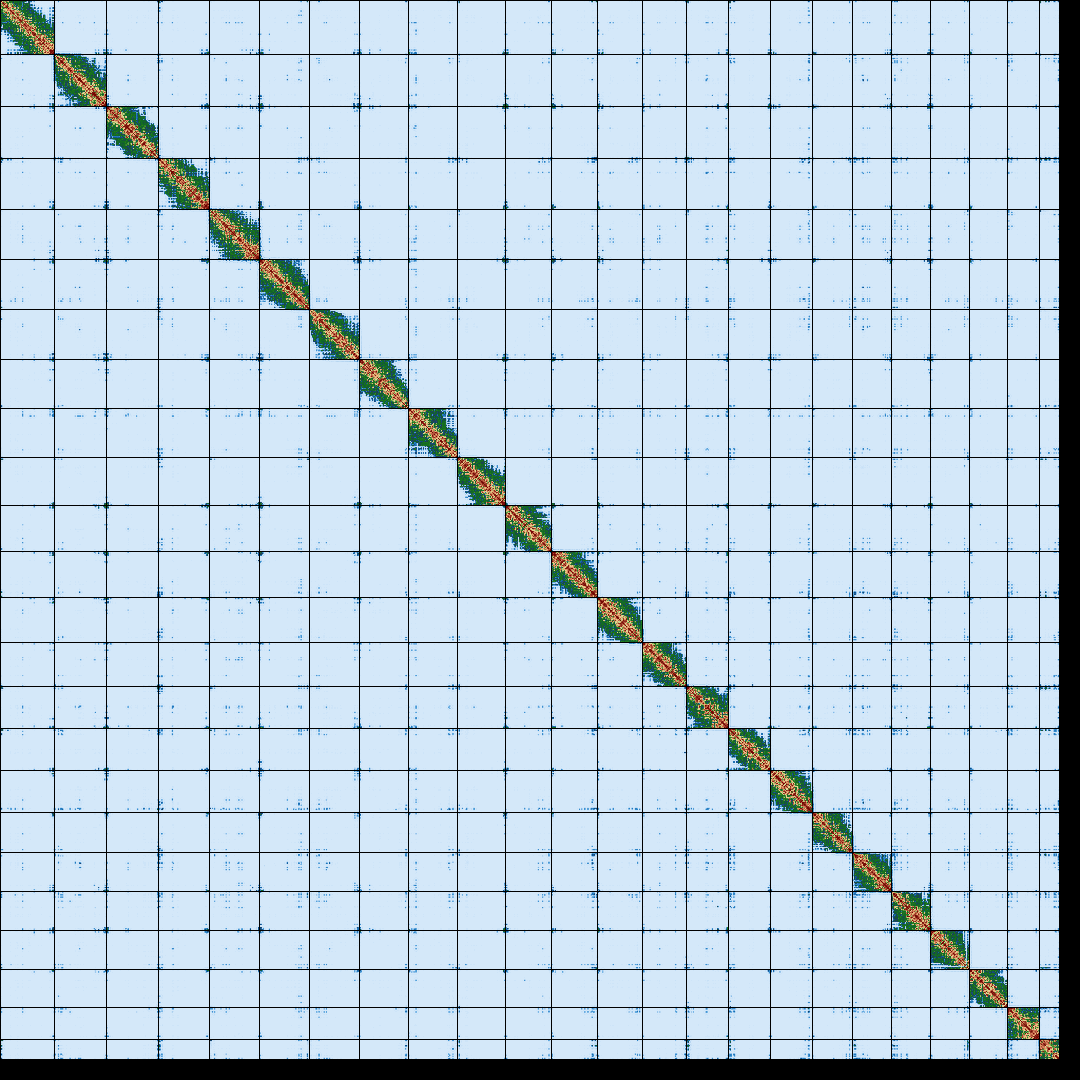
relatively low cost. With long reads and with the help of Hi-C, researchers can generate chromosome
level assemblies without the need for complex computational algorithms or additional sequencing
data. This revolution has transformed the field of genomics and made it possible to
sequence and assemble genomes at an unprecedented pace and scale. This has opened up new avenues of
research in fields such as genetics, medicine, and evolutionary biology.
SeBiMER has assembled numerous prokaryotic and eukaryotic genomes of divers marine species. Some of these
eukaryotic assemblies where among the most challenging due to high heterozygosity rate and genomic
plasticity like the Pacific oyster, Crassostrea gigas.
SeBiMER is a partner in the PEPR Atlasea project which aims
to sequence 4500 out of 12000 known marine species in the French Exclusive Economic Zone (EEZ) in
metropolitan France and four overseas territories. It's aiming at understanding and analysing ecosystems
that are particularly threatened, fragile, biologically important or economically strategic.
Ongoing and recently published projects